In vitro Micronucleus Test, Chromosomal Aberration Tests, Cytogenetics
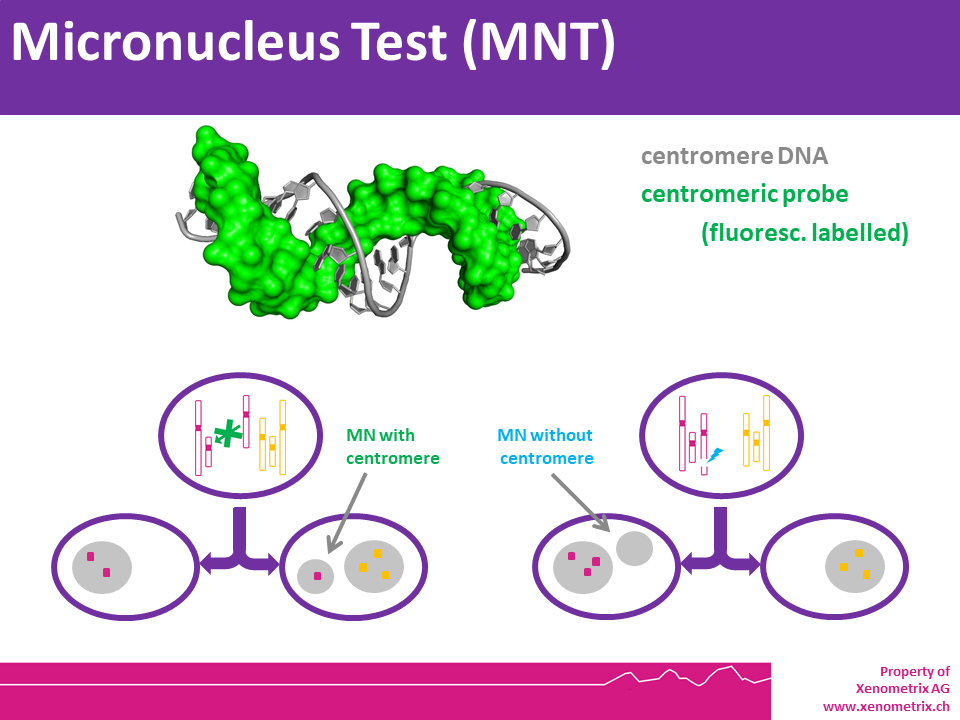
In Vitro MicroNucleus Test MNT (OECD TG487) – Principle
The in vitro MicroNucleus test is one of the most successful and reliable assays for the detection of chemicals with genotoxic potential. Chemicals or physical agents may generate acentric chromosomes (clastogen event) or inappropriately segregate whole chromosomes (aneugen event) leading to genetic damage in the form of an erratic, smaller nucleus called micronucleus.
Both in vitro and in vivo studies can be designed. The former is generally performed using mammalian cell lines, typically upon administration of test items (chemicals or mixtures). In vivo micronucleus assay is performed on peripheral blood cells for cytogenetics research and investigations on the genotoxic effects of radiation and irradiation.
The characteristics of the micronucleus allow to identify the mechanism of action when FISH probes are applied. This information is very important to fully understand the biological details of the genotoxic agent and design follow-up studies to address them.
The Micronucleus assay is described in OECD TG487. Briefly, mammalian cells are grown exponentially during treatment until harvest. Cells must be at various stages of their cell cycles and must undergo at least 1.5-2 cell cycles during exposure with the test chemical. The treatment schedules for different cell lines, primary cell cultures or lymphocytes can vary. Most aneugens and clastogens can be detected during a treatment period of 3 to 6 hours in the absence or presence of metabolic activation by rat liver microsomal fraction S9. After the exposure, medium, S9 (if added) and test compound are removed, fresh media is added and the cells are incubated for a time of 1.5 – 2 cell cycles.
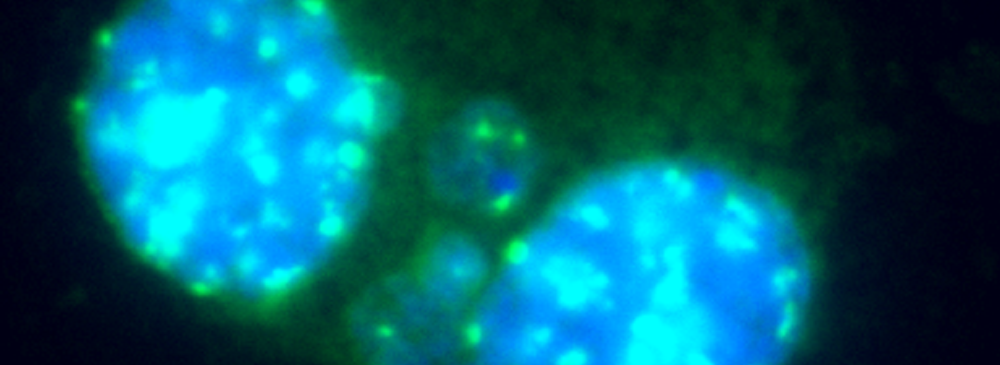
Cell cultures are harvested individually, and cells are prepared for slide preparation (hypotonic treatment or cell spreading). Various techniques can be used to stain the Micronuclei, e.g. Giemsa or specific dyes for staining DNA. Some specific fluorescent dyes can eliminate some of the artifacts associated with non-DNA specific stain: Anti-kinetochore antibodies or FISH with pancentromeric DNA probes together with appropriate DNA counterstaining (DAPI), can be used to identify the contents of micronuclei if mechanistic information is required. Image analysis or flow cytometry can also provide useful information. Please note that chromosome loss is not detected in the chromosome aberration test.
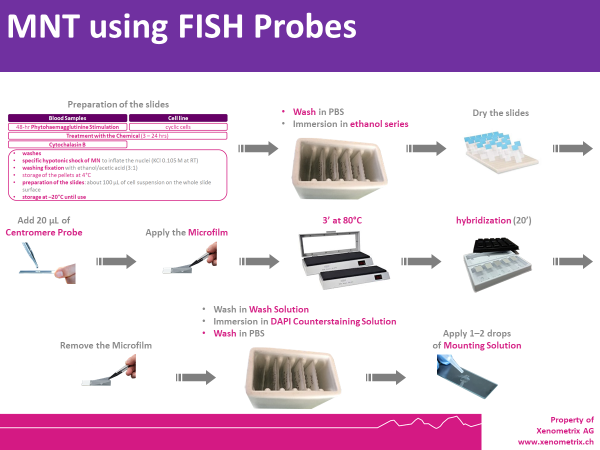
MNT Description using FISH Probes - Rapid MicroNucleus MoA
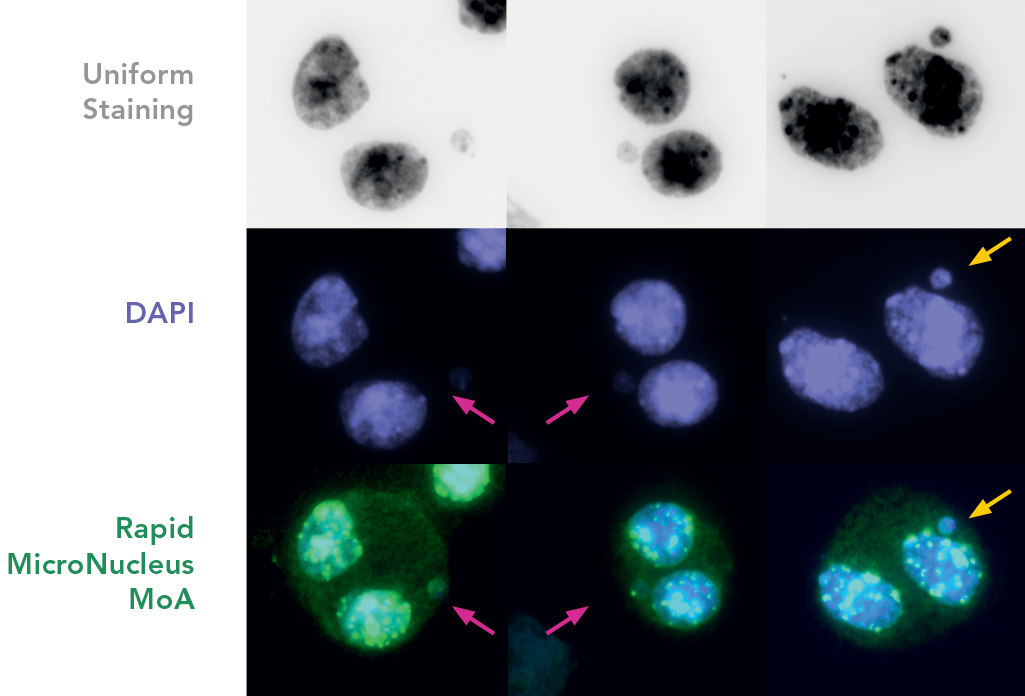
FISH probes and a set of carefully optimized solutions allow to detect centromeres and telomeres in human cells sensitively, using both ex vivo or cell lines, in as fast as 1 hour. A ready to use kit system is available: Rapid Micronucleus MoA
Cells are prepared as per standard protocols according to OECD TG 487 and the slides are sequentially treated with a series of solutions provided with the kit “Rapid Micronucleus MoA”, including a human-specific, highly sensitive centromere probe and a counterstaining solution containing DAPI (4’,6-diamidino-2-phenylindole). In detail, slides are sequentially washed with PBS and an ethanol series after treatment. 20 µL of the centromere probe are then added to the area of interest and the slide is heated at 80°C for 3 minutes and transferred to a humidified chamber for 20 minutes. Washes and a 5-minutes counterstaining step follow prior to the addition of the mounting solution.
Using a 40× magnification objective it will be possible to easily identify micronuclei and assess whether a centromere is present. Automated reading and archiving can be obtained with the slide scanning systems, e.g. Metafer.
Slides are usually ready for evaluation within 1 hour.
Biomonitoring the exposition to low doses of radiation - FISH‑based micronucleus‑centromere assay
FISH probe technology as used in the Rapid Micronucleus MoA Kit can also be used in cytogenetic analysis, in addition to physical dosimetry, i.e. people exposed in medical or natural radiation.
More information.
Rapid MicroNucleus MoA – References
- Natarajan AT, Boei JJ, Darroudi F, Van Diemen PC, Dulout F, Hande MP and Ramalho AT. 1996. Current cytogenetic methods for detecting exposure and effects of mutagens and carcinogens. Environ Health Perspect. 104(Suppl 3):445–448.
- Thierens H, Vral A, Morthier R, Aousalah B and De Ridder L. 2000. Cytogenetic monitoring of hospital workers occupationally exposed to ionizing radiation using the micronucleus centromere assay. Mutagenesis. 15:245–249.
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD and Thomas P. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 26(1):125–132.
- Vral A, Decorte V, Depuydt J, Wambersie A, Thierens H. 2016. A semi-automated FISH-based micronucleus-centromere assay for biomonitoring of hospital workers exposed to low doses of ionizing radiation. Mol Med Rep. 14:103–10.
- Cuceu C, Colicchio B, Jeandidier E, Junker S, Plassa F, Shim G, Mika J, Frenzel M, Al Jawhari M., Hempel WM, O'Brien G, Lenain A, Morat L, Girinsky T, Dieterlen A, Polanska J, Badie C, Carde P, M'Kacher R. 2018. Independent mechanisms lead to genomic instability in Hodgkin lymphoma: Microsatellite or chromosomal instability. Cancers (Basel). 10(7).pii:E233. Erratum in: 2019. Cancers (Basel). 11(6).pii:E757.
- Zaguia N., Laplagne E., Colicchio B., Cariou O., Al Jawhari M., Heidingsfelder L., Hempel WM, Bel Hadj Jrad B, Jeandidier E, Dieterlen A, Carde P, Voisin P, M’Kacher R. 2020. A new tool for genotoxic risk assessment: Reevaluation of the cytokinesis block micronucleus assay using semi-automated scoring following telomere and centromere staining. Mutat Res Gen Tox En. 850-851:503143.

 Navigation
Navigation